Nathi Med |Indlela ye-multi-omics yokwenza imephu yesimila esihlanganisiwe, isimo somzimba sokuzivikela kanye nesandulela ngculazi somdlavuza we-colorectal kuveza ukusebenzisana kwe-microbiome namasosha omzimba.
Nakuba izimpawu ze-biomarker zomdlavuza we-colon oyinhloko ziye zacutshungulwa kakhulu eminyakeni yamuva nje, imihlahlandlela yamanje yomtholampilo incike kuphela kusiteji se-tumor-lymph node-metastasis kanye nokutholwa kokukhubazeka kwe-DNA yokulungisa ukungafani (MMR) noma ukungazinzi kwe-microsatellite (MSI) (ngaphezu kokuhlolwa okujwayelekile kwe-pathology. ) ukunquma izincomo zokwelashwa.Abaphenyi baphawule ukuntuleka kokuhlangana phakathi kwezimpendulo zokuzivikela ezisekelwe kufuzo, amaphrofayili e-microbial, kanye ne-tumor stroma ku-Cancer Genome Atlas (TCGA) yomdlavuza we-colorectal cohort kanye nokusinda kwesiguli.
Njengoba ucwaningo luqhubeka, izici zobuningi bomdlavuza oyinhloko we-colorectal, okuhlanganisa amangqamuzana omdlavuza, i-immune, i-stromal, noma i-microbial nature yomdlavuza, kuye kwabikwa ukuthi kuhlobana kakhulu nemiphumela yomtholampilo, kodwa kusenokuqonda okulinganiselwe kokuthi ukusebenzisana kwabo kuthinta kanjani imiphumela yesiguli. .
Ukwehlukanisa ubudlelwano phakathi kobunzima be-phenotypic kanye nomphumela, ithimba labacwaningi abavela eSidra Institute of Medical Research eQatar muva nje lenze futhi laqinisekisa amaphuzu ahlanganisiwe (mICRoScore) ahlonza iqembu leziguli ezinamazinga amahle okusinda ngokuhlanganisa izici ze-microbiome kanye nokwenqatshwa kwamasosha omzimba. ama-constants (ICR).Ithimba lenze ukuhlaziya okubanzi kofuzo lwamasampula amasha aqandisiwe ezigulini ezingama-348 ezinomdlavuza we-colorectal oyinhloko, okuhlanganisa ukulandelana kwe-RNA kwamathumba kanye nezicubu ezinempilo ezihambisanayo, ukulandelana okuphelele kwe-exome, i-T-cell receptor ejulile kanye nokulandelana kofuzo lwebhaktheriya eyi-16S ye-rRNA, kulekelelwa isimila esiphelele. ukulandelana kwe-genome ukuze kuthuthukiswe i-microbiome.Ucwaningo lwanyatheliswa ku-Nature Medicine ngokuthi "I-tumor ehlanganisiwe, i-immune kanye ne-microbiome atlas yomdlavuza wekoloni".

I-athikili eshicilelwe ku-Nature Medicine
Ukubuka konke kwe-AC-ICAM
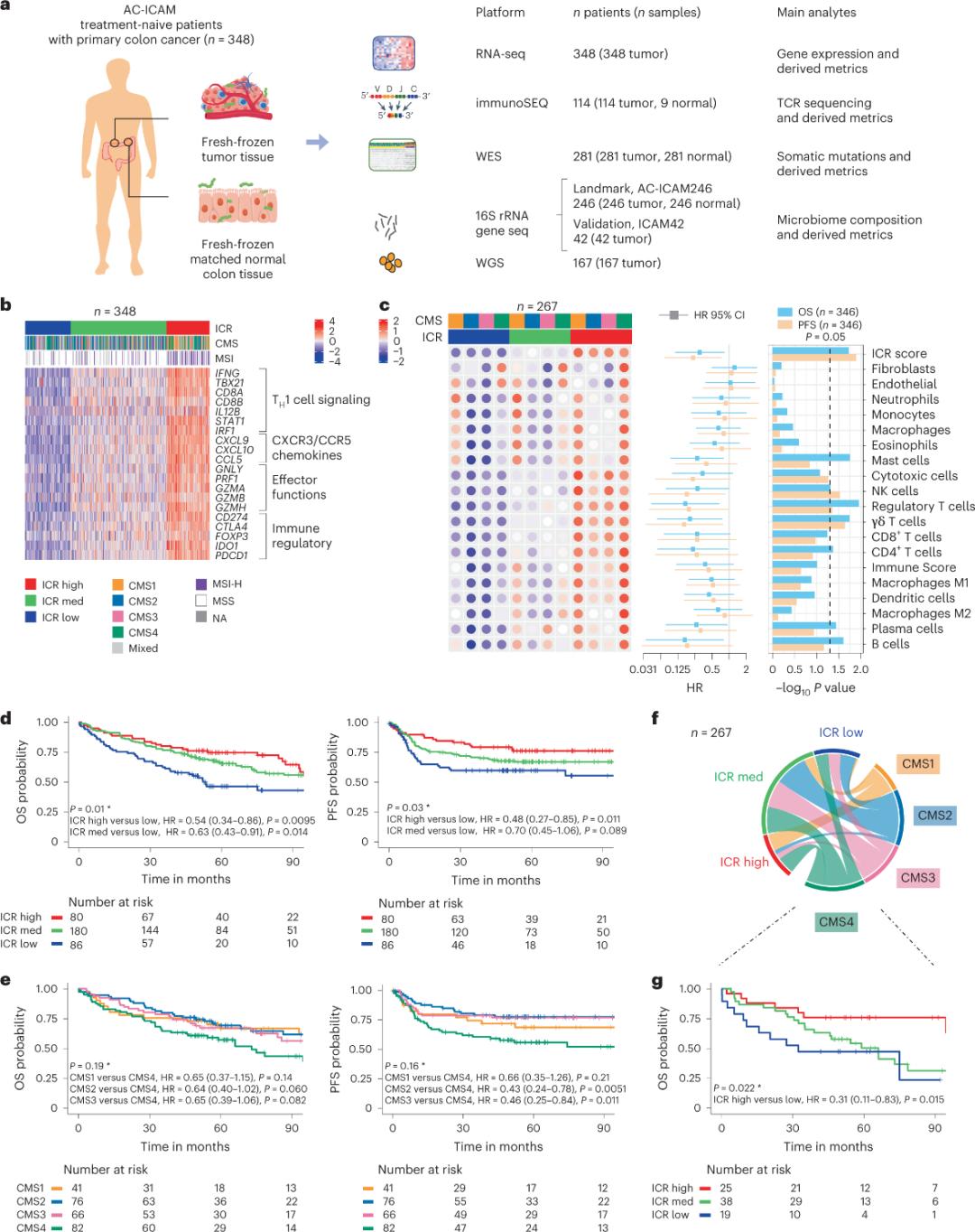
Abacwaningi basebenzise iplathifomu ye-orthogonal genomic ukuhlaziya amasampula amasha aqandiswe isimila futhi bafanise izicubu zekholoni eziseduze ezinempilo (amapheya avamile wesimila) ezigulini ezinokuxilongwa kwe-histologic yomdlavuza wekoloni ngaphandle kokwelashwa kwe-systemic.Ngokusekelwe ku-whole-exome sequencing (WES), ukulawulwa kwekhwalithi yedatha ye-RNA-seq, nokuhlolwa kwezinqubo zokufakwa, idatha ye-genomic evela ezigulini ze-348 yagcinwa futhi yasetshenziselwa ukuhlaziywa komfula ngokulandela okuphakathi kweminyaka engu-4.6.Ithimba labacwaningi liqambe le nsiza ngokuthi i-Sidra-LUMC AC-ICAM: Imephu kanye nomhlahlandlela wokusebenzisana kwe-immune-cancer-microbiome (Umfanekiso 1).
Ukuhlukaniswa kwamangqamuzana kusetshenziswa i-ICR
Ukuthwebula iqoqo lezimpawu zofuzo ezijwayelekile zokuvikela ukuzivikela komdlavuza, okubizwa ngokuthi yi-immune constant of rejection (ICR), ithimba labacwaningi lenze i-ICR ibe ngcono ngokuyihlanganisa ibe yiphaneli yezakhi zofuzo ezingama-20 ehlanganisa izinhlobo ezahlukene zomdlavuza, okubandakanya i-melanoma, umdlavuza wesinye, kanye nomdlavuza wesinye. umdlavuza webele.I-ICR ibuye yahlotshaniswa nokuphendula kwe-immunotherapy ezinhlobonhlobo zezinhlobo zomdlavuza, kuhlanganise nomdlavuza webele.
Okokuqala, abacwaningi baqinisekisa isiginesha ye-ICR yeqembu le-AC-ICAM, besebenzisa indlela yokuhlukanisa izakhi zofuzo ye-ICR ukuze bahlukanise iqoqo libe ngamaqoqo amathathu/ama-subtypes amasosha omzimba: i-ICR ephezulu (izimila ezishisayo), i-ICR emaphakathi ne-ICR ephansi (ebandayo. izimila) (Umfanekiso 1b).Abacwaningi babonise ukuthambekela kokuzivikela komzimba okuhlotshaniswa ne-consuss molecular subtypes (CMS), ukuhlukaniswa okusekelwe ku-transcriptome yomdlavuza wekoloni.izigaba ze-CMS zazihlanganisa i-CMS1/immune, CMS2/canonical, CMS3/metabolic kanye ne-CMS4/mesenchymal.Ukuhlaziywa kubonise ukuthi izikolo ze-ICR zazihlotshaniswa kabi nezindlela ezithile zamangqamuzana omdlavuza kuwo wonke ama-subtypes e-CMS, futhi ukuhlobana okuhle ne-immunosuppressive kanye nezindlela ezihlobene ne-stromal kwabonwa kuphela kumathumba e-CMS4.
Kuwo wonke ama-CMS, ukuchichima kwe-cell killer (NK) cell kanye ne-T cell subsets kwakuphakeme kakhulu ku-ICR high immune subtypes, nokuhlukahluka okukhulu kwamanye ama-leukocyte subsets (Umfanekiso 1c) .I-ICR immune subtypes yayine-OS ehlukile ne-PFS, ngokukhula okuqhubekayo ku-ICR ukusuka phansi kuye phezulu (Umfanekiso 1d), oqinisekisa indima yokubikezela ye-ICR kumdlavuza we-colorectal.
Umfanekiso 1. Idizayini yocwaningo lwe-AC-ICAM, isignesha yofuzo ehlobene nokuzivikela komzimba, ama-immune and molecular subtypes kanye nokusinda.
I-ICR ithwebula amaseli e-T athuthukiswe isimila, akhuliswe ngokuhlanganyela
Idlanzana kuphela lamaseli e-T afaka izicubu zesimila okubikwe ukuthi aqondene ngqo nama-antigen wesimila (ngaphansi kuka-10%).Ngakho-ke, iningi lamaseli e-intra-tumor T abizwa ngokuthi ama-T cell abukele (ama-T cell abukele).Ukuhlobana okuqine kakhulu nenani lamaseli e-T avamile anama-TCR akhiqizayo abonwa ku-stromal cell kanye ne-leukocyte subpopulations (etholwe yi-RNA-seq), engasetshenziswa ukulinganisa ukugcwala kwamaseli e-T (Umfanekiso 2a).Kumaqoqo e-ICR (sekukonke kanye nesigaba se-CMS), ukuhlangana okuphezulu kakhulu kwe-immune SEQ TCRs kwabonwa ku-ICR-high kanye ne-CMS subtype CMS1/amaqembu amasosha omzimba (Umfanekiso 2c), anengxenye ephezulu kakhulu yamathumba aphezulu e-ICR.Ukusebenzisa i-transcriptome yonke (izakhi zofuzo ze-18,270), izakhi zofuzo ze-ICR eziyisithupha (IFNG, STAT1, IRF1, CCL5, GZMA, ne-CXCL10) zaziphakathi kwezakhi zofuzo eziyishumi eziphezulu ezihlotshaniswa kahle ne-TCR immune SEQ clonality (Figure 2d).I-ImmunoSEQ TCR clonality ihlotshaniswe ngokuqinile nezakhi zofuzo eziningi ze-ICR kunokuxhumana okubonwa kusetshenziswa omaka be-CD8+ abasabela ngesimila (Umfanekiso 2f no-2g).Sengiphetha, ukuhlaziya okungenhla kuphakamisa ukuthi isiginesha ye-ICR ithwebula ubukhona bamaseli e-T athuthukisiwe, akhuliswe ngokufana futhi ingase ichaze imithelela yawo yokubikezela.
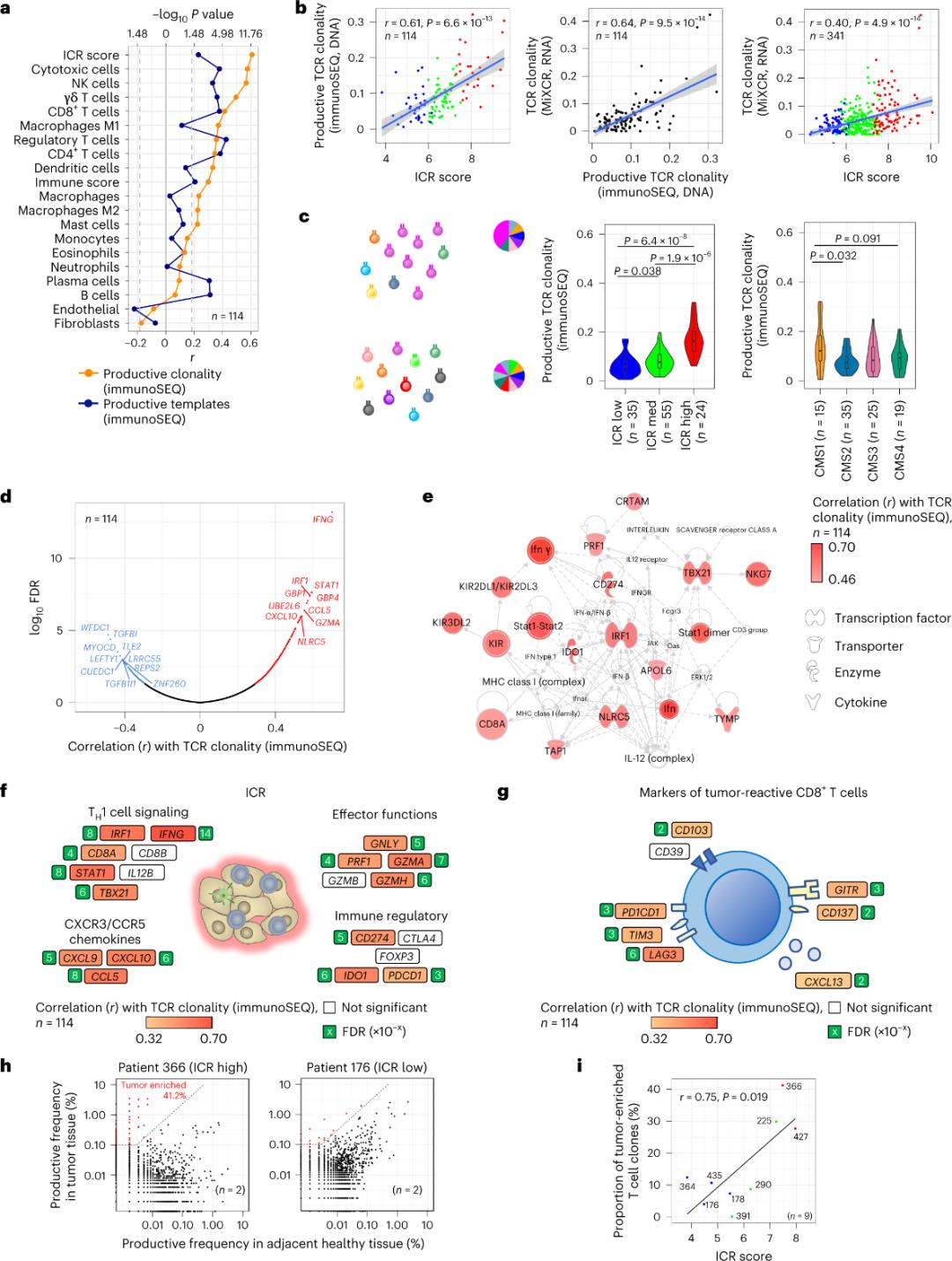
Umfanekiso we-2. Amamethrikhi we-TCR nokuhlobana nezakhi zofuzo ezihlobene nokuzivikela komzimba, ama-immune and molecular subtypes.
Ukwakheka kwe-Microbiome ezicutshini zomdlavuza we-colon onempilo
Abacwaningi benze ukulandelana kwe-16S rRNA besebenzisa i-DNA ekhishwe esimila esifaniswayo kanye nezicubu zekholoni ezinempilo ezigulini ezingama-246 (Umfanekiso 3a).Ukuze kuqinisekiswe, abacwaningi baphinde bahlaziya idatha yokulandelana kofuzo lwe-16S rRNA kusuka kumasampuli engeziwe angama-42 angenayo i-DNA evamile etholakalayo ukuze ihlaziywe.Okokuqala, abacwaningi baqhathanise ubuningi bezitshalo phakathi kwamathumba afanayo kanye nezicubu zekholoni ezinempilo.I-Clostridium perfringens yanda kakhulu emathunjini uma kuqhathaniswa namasampula anempilo (Umfanekiso 3a-3d).Kwakungekho mehluko obalulekile ezinhlobonhlobo ze-alpha (ukwehlukahlukana nobuningi bezinhlobo zezilwane kusampula eyodwa) phakathi kwamasampula ethumba namasampula anempilo, futhi ukuncipha okuncane kokuhlukahluka kwamagciwane kubonwe kumathumba aphezulu e-ICR ahlobene nezimila eziphansi ze-ICR.
Ukuze kutholwe izinhlangano ezifanele ngokomtholampilo phakathi kwamaphrofayili e-microbial kanye nemiphumela yomtholampilo, abacwaningi bahlose ukusebenzisa idatha yokulandelana kofuzo lwe-16S rRNA ukuhlonza izici ze-microbiome ezibikezela ukusinda.Ku-AC-ICAM246, abacwaningi basebenzise imodeli yokuhlehla ye-OS Cox ekhethe izici ezingu-41 ezinama-coefficients angewona aziro (ahambisana nengozi yokufa ehlukile), ebizwa ngokuthi i-MBR classifiers (Figure 3f).
Kuleli qembu lokuqeqesha (ICAM246), isikolo esiphansi se-MBR (MBR<0, i-MBR ephansi) sasihlotshaniswa nengozi ephansi kakhulu yokufa (85%).Abacwaningi baqinisekise ukuhlangana phakathi kwe-MBR ephansi (ingozi) ne-OS enwetshiwe kumaqoqo amabili aqinisekisiwe ngokuzimela (ICAM42 kanye ne-TCGA-COAD).(Umfanekiso wesi-3) Ucwaningo lubonise ukuhlobana okuqinile phakathi kwe-endogastric cocci kanye nezikolo ze-MBR, ezazifana ne-tumor kanye nezicubu ze-colon enempilo.
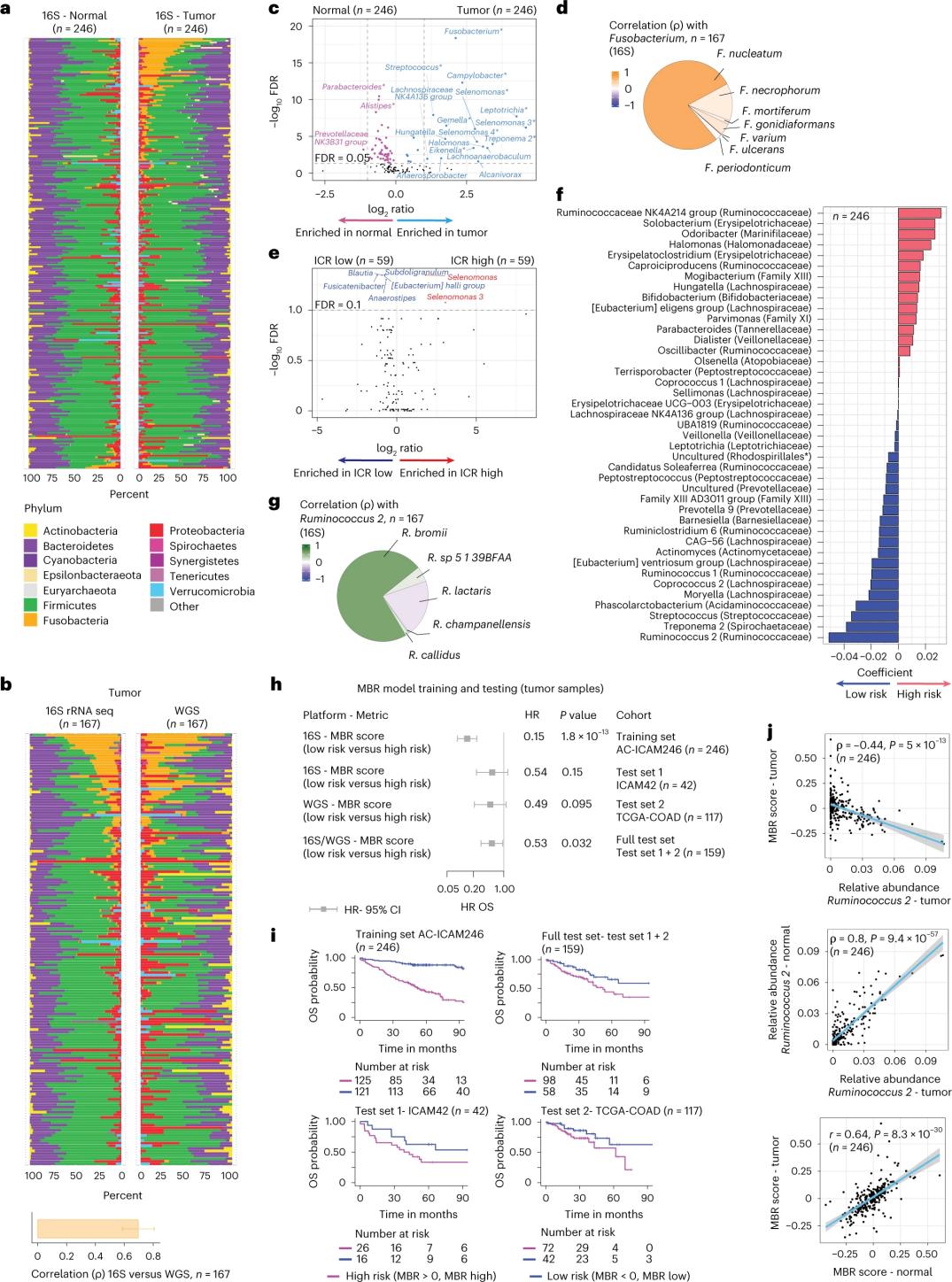
Umfanekiso 3. I-Microbiome ku-tumor kanye nezicubu ezinempilo kanye nobuhlobo ne-ICR kanye nokusinda kwesiguli.
Isiphetho
Indlela ye-multi-omics esetshenziswe kulolu cwaningo ivumela ukutholwa okuphelele nokuhlaziywa kwesiginesha yamangqamuzana empendulo yokuzivikela komzimba kumdlavuza we-colorectal futhi iveze ukusebenzisana phakathi kwe-microbiome nesistimu yomzimba.Ukulandelana kwe-TCR okujulile kwesimila kanye nezicubu ezinempilo kwembula ukuthi umphumela wokubikezela we-ICR ungase ube ngenxa yekhono layo lokuthwebula ama-cell clones athuthukiswe isimila futhi mhlawumbe ne-tumor antigen-specific T cell.
Ngokuhlaziya ukwakheka kwe-tumor microbiome kusetshenziswa ukulandelana kofuzo kwe-16S rRNA kumasampuli e-AC-ICAM, ithimba lihlonze isiginesha ye-microbiome (isikolo sengozi ye-MBR) enevelu eqinile yokubikezela.Nakuba lesi siginesha sithathwe kumasampula wesimila, kube nokuhlobana okuqinile phakathi kwe-colorectum enempilo ne-tumor MBR score, okuphakamisa ukuthi lesi siginesha singathwebula ukwakheka kwe-gut microbiome yeziguli.Ngokuhlanganisa izikolo ze-ICR ne-MBR, bekungenzeka ukuhlonza nokuqinisekisa i-biomarker yomfundi we-multi-omic ebikezela ukusinda ezigulini ezinomdlavuza wekoloni.Idathasethi yocwaningo ene-multi-omic ihlinzeka ngesisetshenziswa sokuqonda kangcono ibhayoloji yomdlavuza wamathumbu futhi isize ukuthola izindlela zokwelapha eziqondene nawe.
Isikhathi sokuthumela: Jun-15-2023
 中文网站
中文网站